Abstract
Background
Next-generation sequencing (NGS) based on liquid biopsy has been an emerging technology to identify tumor-specific genetic aberrations in adult lymphoma. However, there were few studies on the genomic profiling of plasma circulating tumor-derived DNA (ctDNA) in pediatric mature B-NHL.
Methods
Paraffin-embedded tissue (FFPE) and plasma samples from the newly diagnosed patients were collected and sequenced by 475 genes panel before, during and post of treatment. Clinical stage system, risk stratification and treatment of pediatric mature B-NHL followed a the modified BFM-95 protocol.
Results
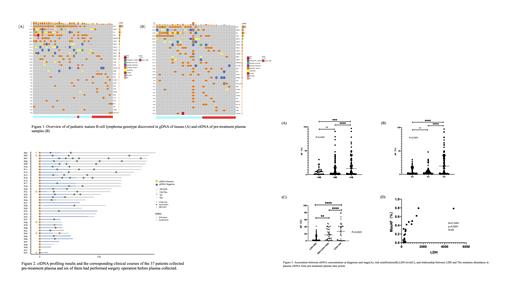
A total of 53 pediatric mature B-NHLs were enrolled,including 35 Burkitt lymphoma, 18 diffused large B cell lymphoma/high-grade B cell lymphoma (DLBCL/HGBCL). We collected 38 tissue and 124 plasma samples for somatic mutation testing. The number of somatic mutations and the TOP 5 genes detected in the 38 tissues and 31 baseline plasma samples, were 416 vs 496, and MYC(71%), DDX3X(45%),ID3 (42%), TP53(40%), SMARCA4(29%) (Fig1. A)vs MYC(52%), DDX3X(45%),ID3(42%), TP53(36%), GNA13(23%) (Fig1. B), respectively. The median allele frequency of mutations in plasma was 3%(ranged from 0.2% to 96.6%) and MYC, DDX3X, ID3, TP53, SMARCA4,ARID1A shows higher max somatic allele frequency (MSAF), indicated that was the early events in tumor genesis.
The sensitivity of plasma ctDNA to detecting tissue mutations was 63.4% in the 19 matched samples (11 samples from BL and 8 samples from DLBCL/HGBCL) and the sensitivity in BL and DLBCL/HGBCL were 64.1% and 62%, respectively. All genomic alteration types, including single nucleotide variants (SNVs), indels, and gene fusions were detected in similar proportions in each sample type and the gene mutation rate of every gene detected in paired tissue and ctDNA samples.
Among the 37 mature B-NHL patients, 6 patients were collected plasma samples after resection of tumor, of which 4 patients was not detected the somatic gene mutations in plasma (Fig. 2). The abundance of ctDNA in patients with stage IV (N=10) was significantly higher than that of stage I-II (N=6) (P=0.0002) and stage III patients(N=15)(P<0.0001)(Fig3. A). Similarly the abundance of ctDNA mutations in high risk patients(N =11)was significantly higher than that of low risk patients (N=8)(P<0.0001) and Medium risk patients (N=12) (P<0.0001) (Fig3. B). MSAF of ctDNA mutations was significantly correlated with LDH and the abundance of ctDNA mutations (P<0.0001) (Fig3. C,D).
With a median follow-up of 182 Days, 33 patients have completed anti-tumor treatment, 27 patients completed post-treatment PET-CT, and 20 patients have done ctDNA testing synchronously. PET-CT showed tumor residue in 4 patients, of which 2 patients showed no tumor residue in pathology and ctDNA, 1 patient showed tumor residue in pathology but not in ctDNA and with tumor progression 6 months after treatment, 1 patient unable to take biopsy showed no tumor residue in ctDNA and was no tumor reccourence with regular follow-up. At the last follow-up, 1 patient was disease progression, and all of the 53 patients survived.
Conclusion
Plasma ctDNA testing by NGS was practicable in pediatric mature B-NHL. The abundance of ctDNA is significantly related to tumor burden. CtDNA testing may be more sensitive than PET-CT for residual disease assessment. Nevertheless, sample size expansion is required to verify such conclusions.
No relevant conflicts of interest to declare.